Rag-and-bone science: interesting results from recycled data
I was surprised and thrilled to learn that I was a joint winner of Parasitology Journal’s Early Career Research Award 2024 for my paper “Revisiting fecal metatranscriptomics analyses of macaques with idiopathic chronic diarrhoea with a focus on trichomonad parasites”. This work arose from a need to find new ways of generating interesting results for my PhD thesis while I was unable to access the lab during the Covid-19 lockdowns. The idea was to leverage the vast wealth of next-generation sequencing datasets available in online databases to address new biological questions.
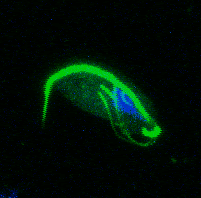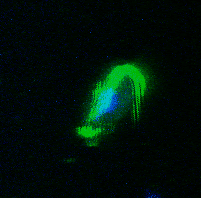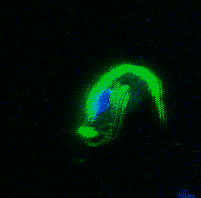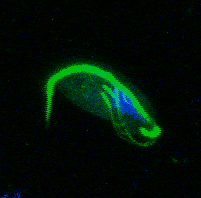
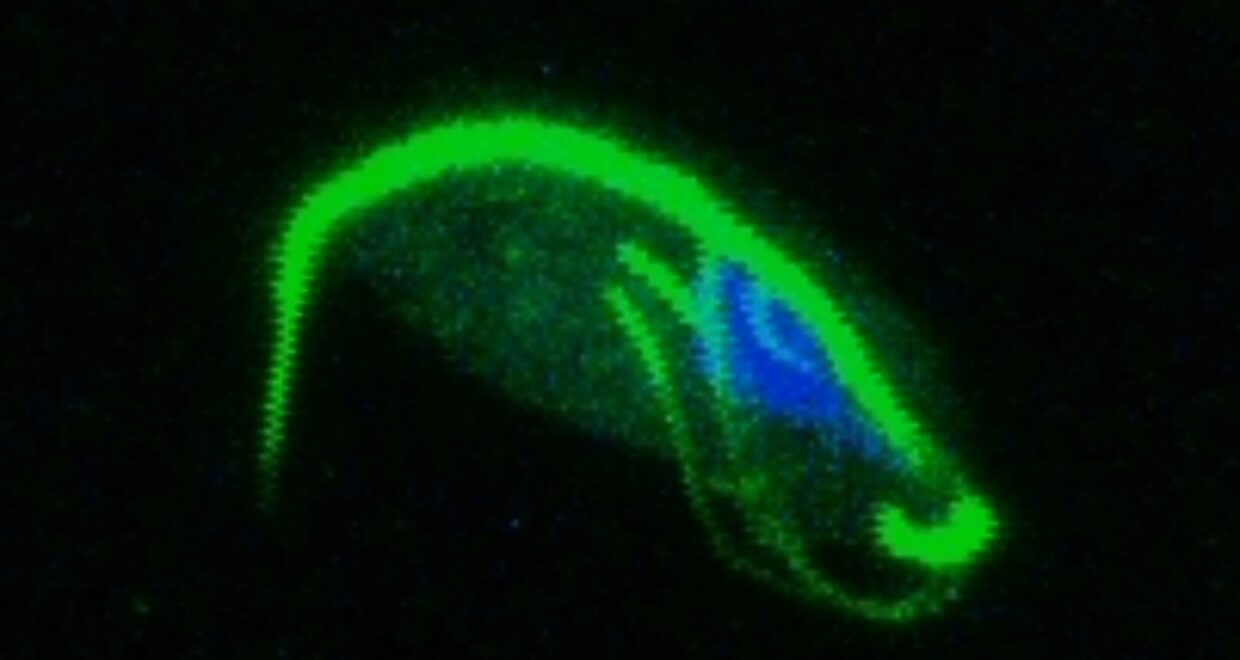
My PhD focused on investigating how Trichomonads have evolved to infect the mucosal surfaces of such a wide range of animal hosts. Trichomonads are an unusual group of highly diverged microbial eukaryotes, which ferment carbon sources to produce hydrogen in their reductively evolved mitochondrion, called the hydrogenosome. The group includes species of major medical and ecological importance. Trichomonas vaginalis is the most common non-viral STD in the world, and is associated with urogenital cancers and increased HIV transmission. Trichomonas gallinae is an avian parasite which was responsible for the early 2000s outbreak in European finches, resulting in dramatic population declines. Importantly, these two species share a recent avian-infecting ancestor, highlighting their evolutionary plasticity to switch between hosts.
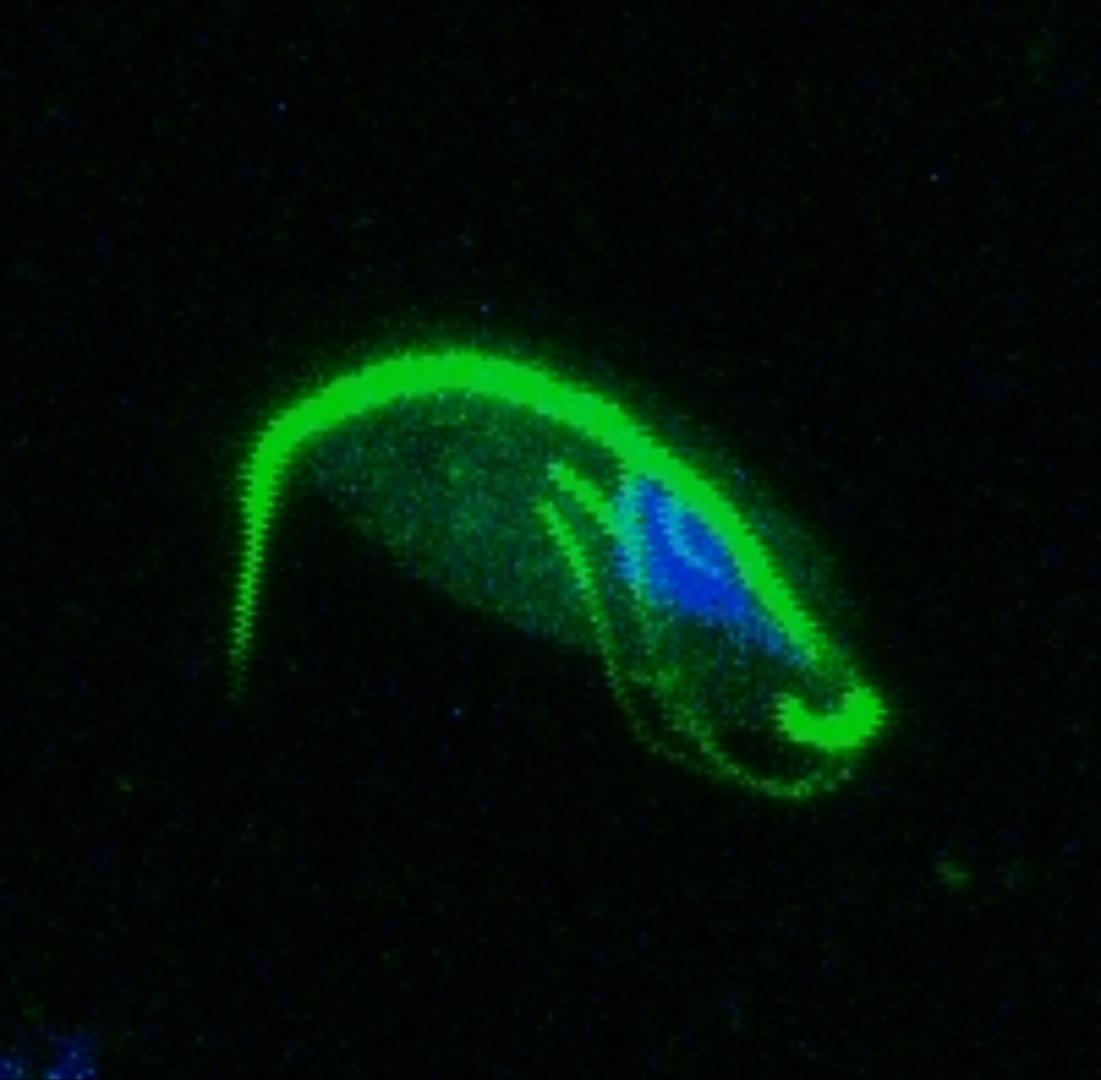
A study by Westreich and colleagues used fecal metatranscriptomics to investigate chronic idiopathic diarrhoea in laboratory rhesus macaques. The authors reported T. vaginalis-like sequences in the dataset, indicating the presence of a related parasite. We took this serendipitous opportunity to investigate the parasite’s transcriptional behaviour during in vivo infection, which was a first for Trichomonad research. This reverse approach to research required me to consider as many ways as possible to exploit the existing dataset, rather than designing an experiment to address a specific question. Assembled transcript sequences from the parasites allowed us to identity them phylogenetically. Unexpectedly, this revealed the presence of three separate Trichomonad genera, two of which hadn’t been previously reported in macaques. Other assembled transcripts from the parasites revealed the genes that they were expressing. This validated the in vivo importance of metabolic pathways which had previously been identified in vitro in T. vaginalis, including those potentially involved in utilising arginine and bacterial cells as nutrient sources. Previous work had shown the importance of trichomonad-bacteria interactions for disease outcome and parasite growth. We investigated this by testing for correlations between bacterial and parasite transcript abundances. The results suggested that Tetratrichomonas occupied a central position in a network of correlated bacterial species. The association we identified between Tetratrichomonas and Prevotella was particularly fascinating; a similar correlation between T. vaginalis and Prevotella is associated with bacterial vaginosis in humans. My more recent re-analysis of 16S rRNA profiling data from pigeons also highlighted an association between T. gallinae and Prevotella. This suggests that specific parasite-bacteria interactions could be highly conserved across different parasite and host species.
This work paved the way for my ongoing career in bioinformatics. A similar rag-and-bone man approach to research, recycling existing sequencing data to address new questions, has allowed me to make progress regardless of success in the lab. I’d like to thank my PhD supervisor Robert Hirt, Westreich and colleagues who generated the original data that made this work possible, and the BBSRC for funding my PhD project. Only researchers who are no more than 7 years post award of their PhD were eligible to receive the award. Decisions on the winning papers were made by Russell Stothard and his team of supporting Editors. The winning papers are freely available.
Only researchers who are no more than 7 years post award of their PhD were eligible to receive the award. Decisions on the winning papers were made by Russell Stothard and his team of supporting Editors. The winning papers are freely available. Dr. Bailey’s winning paper is available to read here: Revisiting fecal metatranscriptomics analyses of macaques with idiopathic chronic diarrhoea with a focus on trichomonad parasites